Palmer penguins
This page is a work-in-progress.
The Palmer penguins dataset is great for practising R!
This package contains data on 3 species of penguins, collected from 3 islands in the Palmer Archipelago, Antarctica. A total of 344 penguins.
library(palmerpenguins)
dim(penguins) #data set with 344 rows & 8 columns#> [1] 333 8head(penguins)#> # A tibble: 6 × 8
#> species island bill_length_mm bill_depth_mm flipper_length_mm
#> <fct> <fct> <dbl> <dbl> <int>
#> 1 Adelie Torgersen 39.1 18.7 181
#> 2 Adelie Torgersen 39.5 17.4 186
#> 3 Adelie Torgersen 40.3 18 195
#> 4 Adelie Torgersen 36.7 19.3 193
#> 5 Adelie Torgersen 39.3 20.6 190
#> 6 Adelie Torgersen 38.9 17.8 181
#> # ℹ 3 more variables: body_mass_g <int>, sex <fct>, year <int>#> [1] 333 8Plot of numeric X & Y parameters
Plot bill_length_mm vs bill_length_mm, bill_depth_mm or bill_length_mm, and fill colours by a third numeric variable bill_length_mm.
We’ll split them by species and sex with facet_grid(sex ~ species), and fit straight lines through these variables with geom_smooth(method = "lm") and additional grouping aesthetics as necessary.
plot_xy_CatGroup(data = penguins,
xcol = bill_depth_mm,
ycol = bill_length_mm,
CatGroup = species,
s_alpha = .3,
TextXAngle = 45)+
geom_smooth(method = "lm",
aes(colour = species),
show.legend = FALSE)+
scale_colour_grafify()+
labs(title = "Bill depth vs flipper length")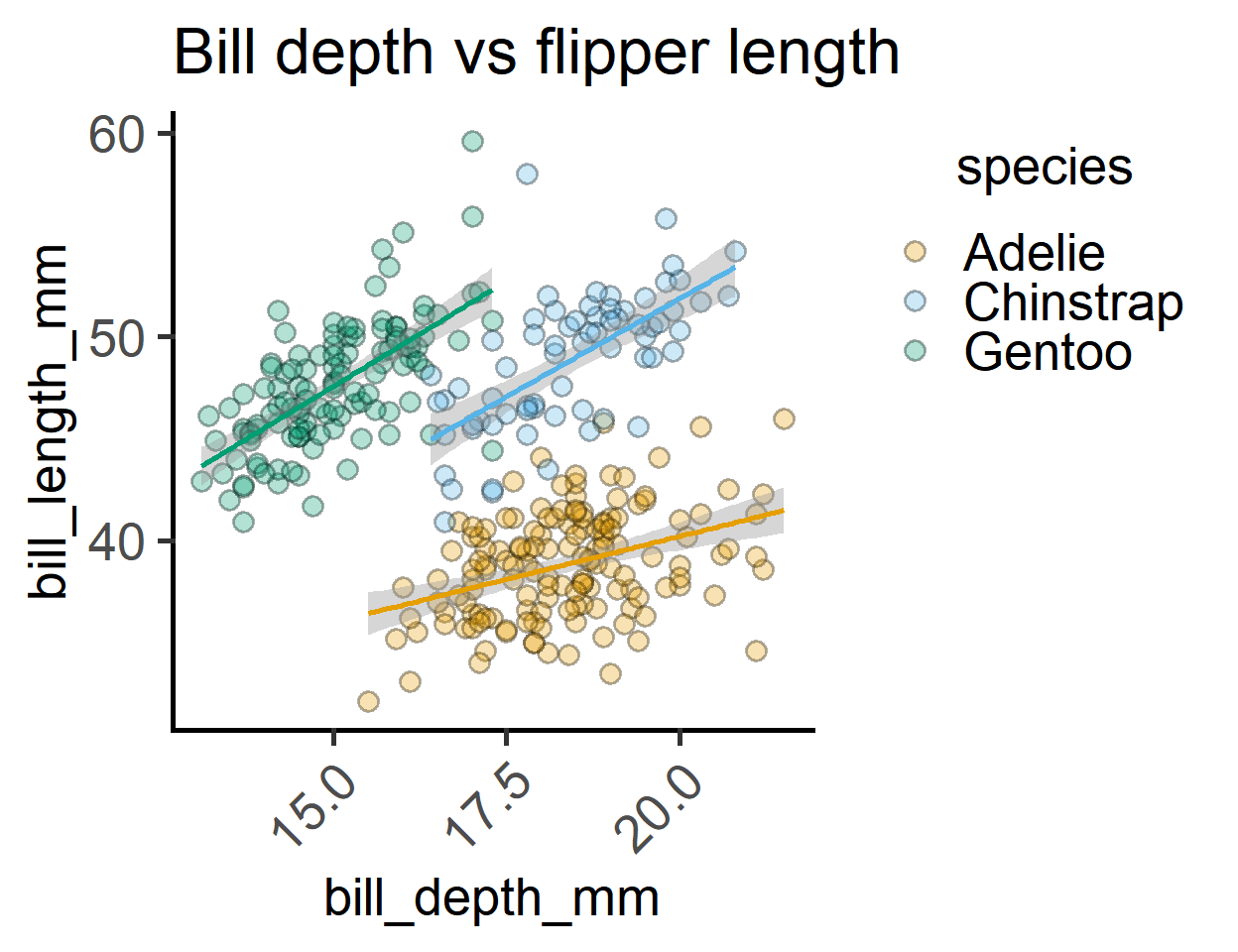
plot_xy_CatGroup(data = penguins,
xcol = bill_depth_mm,
ycol = bill_length_mm,
CatGroup = species,
facet = sex,
s_alpha = .3,
TextXAngle = 45)+
geom_smooth(method = "lm",
aes(colour = species),
show.legend = FALSE)+
scale_colour_grafify()+
labs(title = "Bill depth vs flipper length",
subtitle = "faceted by sex")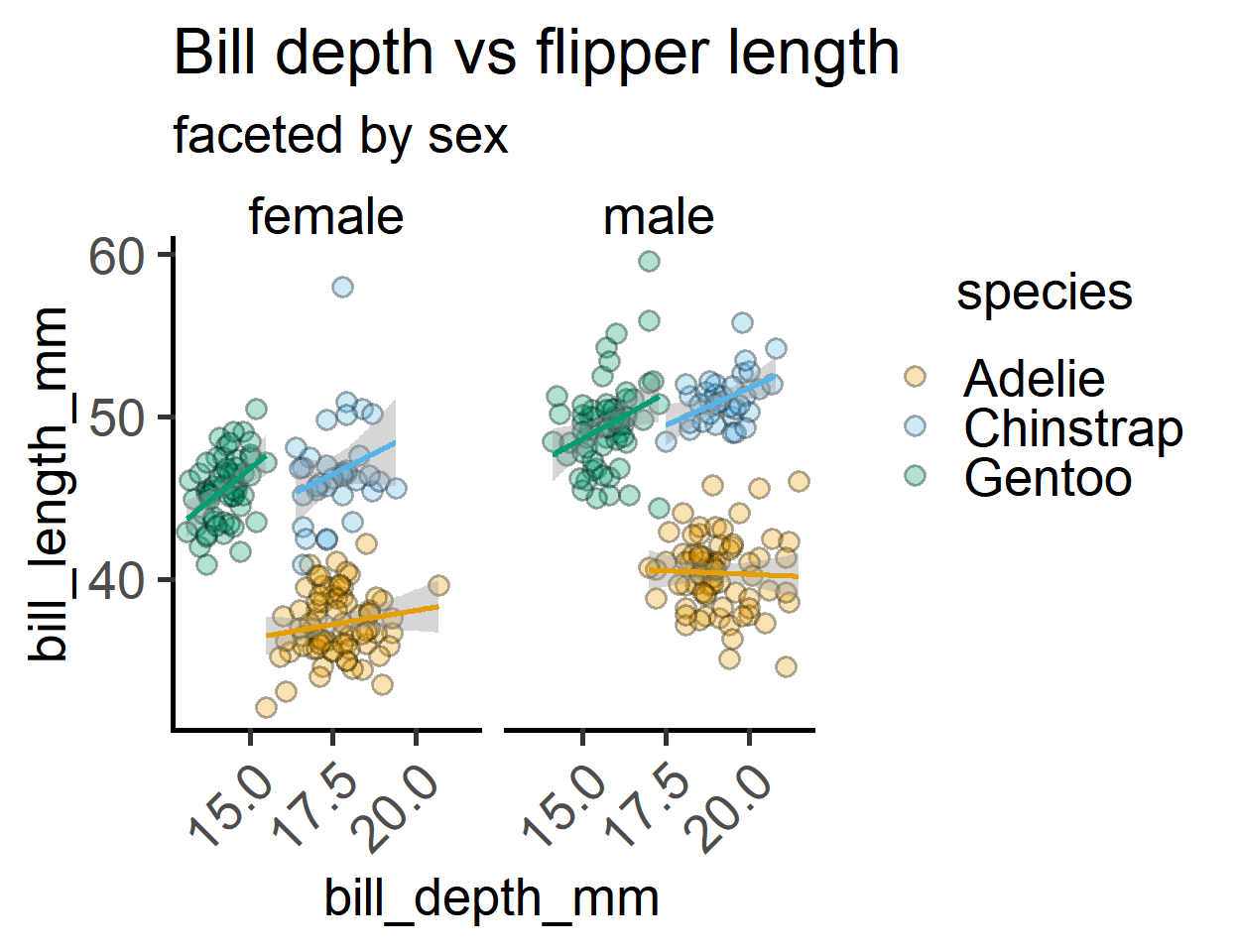
plot_xy_CatGroup(data = penguins,
xcol = bill_depth_mm,
ycol = bill_length_mm,
CatGroup = species,
facet = island,
s_alpha = .3,
TextXAngle = 45)+
geom_smooth(method = "lm",
aes(colour = species),
show.legend = FALSE)+
scale_colour_grafify()+
labs(title = "Bill depth vs flipper length",
subtitle = "faceted by island")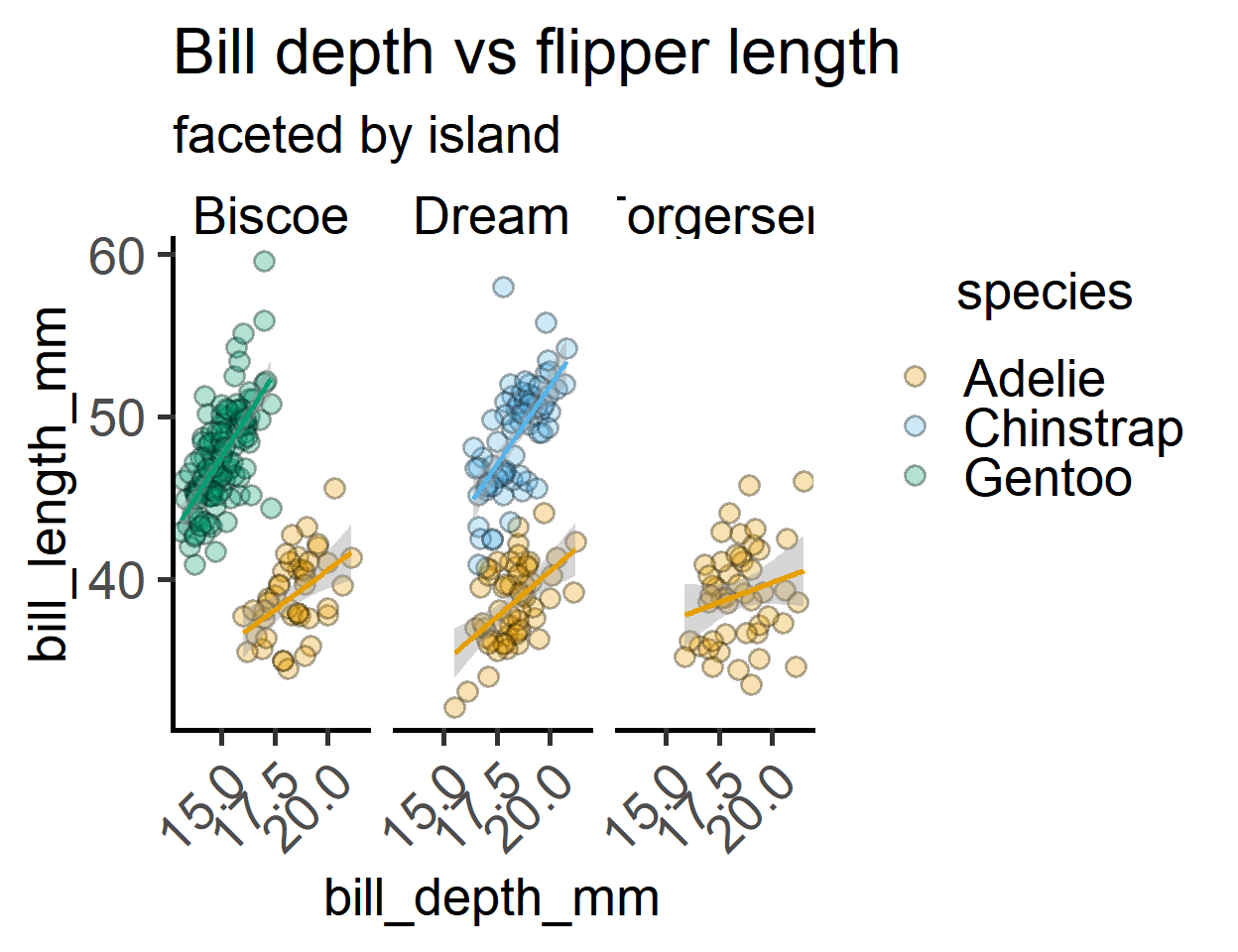
Species & Sex as categorical variables
Plots assessing measures across sex, species & island.
plot_4d_scatterviolin(data = penguins,
xcol = species,
ycol = bill_length_mm,
boxes = sex,
shapes = sex,
facet = island,
jitter = .5,
s_alpha = .1,
v_alpha = .6,
TextXAngle = 45,
bvthick = 0.2)+
labs(title = "Bill length vs species & sex",
subtitle = "faceted by island")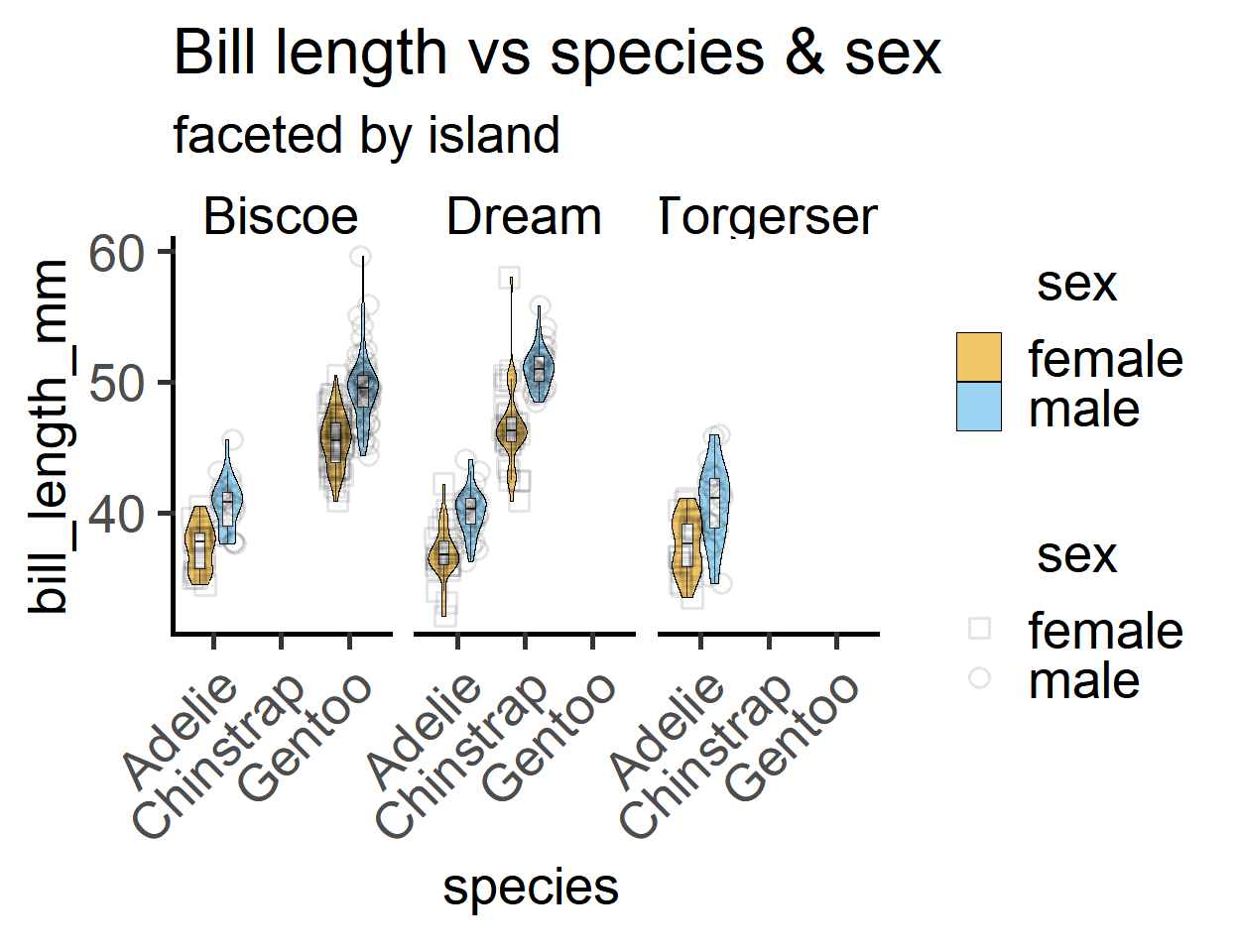
The plot suggests that males have higher body mass than females, and Adelie & Chinstrap males and females are similar in weight, whereas Gentoo penguins have larger body mass.
Facet grid can be used to bring in additional variables.
Linear models
Discreet independent variables sex & species
Let’s assess whether body mass varies by sex and species using linear models for ANOVA.
simple_anova(data = penguins,
Y_value = "bill_length_mm",
c("sex", "species"))#> Anova Table (Type II tests)
#>
#> Response: bill_length_mm
#> Sum Sq Mean sq Df F value Pr(>F)
#> sex 1135.7 1135.7 1 211.8066 <2e-16 ***
#> species 6975.6 3487.8 2 650.4786 <2e-16 ***
#> sex:species 24.5 12.2 2 2.2841 0.1035
#> Residuals 1753.3 5.4 327
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1#save the model
mod1 <- simple_model(data = penguins,
Y_value = "bill_length_mm",
c("sex", "species"))
#model diagnostic with QQ plot
plot_qqmodel(mod1)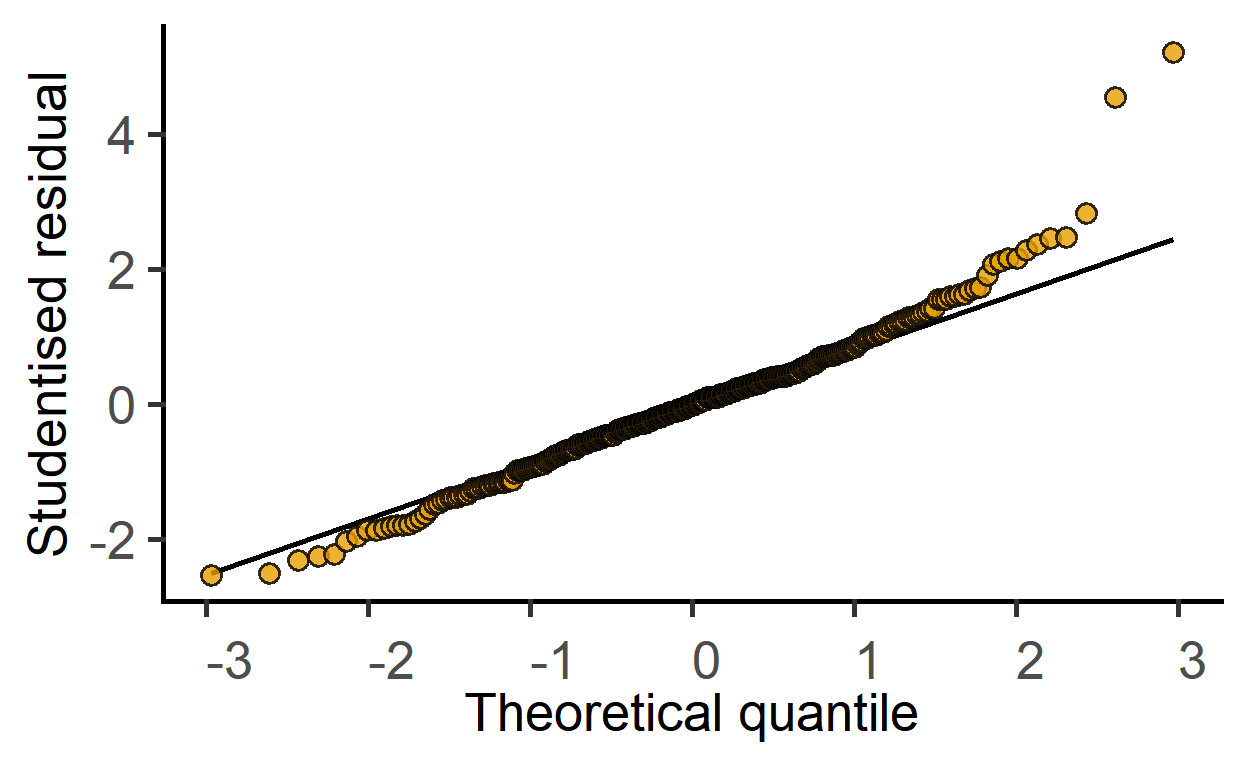
#post-hoc comparisons
posthoc_Levelwise(Model = mod1,
Fixed_Factor = c("sex", "species"))#> $emmeans
#> species = Adelie:
#> sex emmean SE df lower.CL upper.CL
#> female 37.3 0.271 327 36.7 37.8
#> male 40.4 0.271 327 39.9 40.9
#>
#> species = Chinstrap:
#> sex emmean SE df lower.CL upper.CL
#> female 46.6 0.397 327 45.8 47.4
#> male 51.1 0.397 327 50.3 51.9
#>
#> species = Gentoo:
#> sex emmean SE df lower.CL upper.CL
#> female 45.6 0.304 327 45.0 46.2
#> male 49.5 0.296 327 48.9 50.1
#>
#> Confidence level used: 0.95
#>
#> $contrasts
#> species = Adelie:
#> contrast estimate SE df t.ratio p.value
#> female - male -3.13 0.383 327 -8.174 <.0001
#>
#> species = Chinstrap:
#> contrast estimate SE df t.ratio p.value
#> female - male -4.52 0.562 327 -8.049 <.0001
#>
#> species = Gentoo:
#> contrast estimate SE df t.ratio p.value
#> female - male -3.91 0.425 327 -9.207 <.0001posthoc_Levelwise(Model = mod1,
Fixed_Factor = c("species", "sex"))#> $emmeans
#> sex = female:
#> species emmean SE df lower.CL upper.CL
#> Adelie 37.3 0.271 327 36.7 37.8
#> Chinstrap 46.6 0.397 327 45.8 47.4
#> Gentoo 45.6 0.304 327 45.0 46.2
#>
#> sex = male:
#> species emmean SE df lower.CL upper.CL
#> Adelie 40.4 0.271 327 39.9 40.9
#> Chinstrap 51.1 0.397 327 50.3 51.9
#> Gentoo 49.5 0.296 327 48.9 50.1
#>
#> Confidence level used: 0.95
#>
#> $contrasts
#> sex = female:
#> contrast estimate SE df t.ratio p.value
#> Adelie - Chinstrap -9.32 0.481 327 -19.377 <.0001
#> Adelie - Gentoo -8.31 0.407 327 -20.393 <.0001
#> Chinstrap - Gentoo 1.01 0.500 327 2.019 0.0443
#>
#> sex = male:
#> contrast estimate SE df t.ratio p.value
#> Adelie - Chinstrap -10.70 0.481 327 -22.263 <.0001
#> Adelie - Gentoo -9.08 0.402 327 -22.613 <.0001
#> Chinstrap - Gentoo 1.62 0.496 327 3.270 0.0012
#>
#> P value adjustment: fdr method for 3 tests